
What is unusual about the SARS-CoV-2 virus?
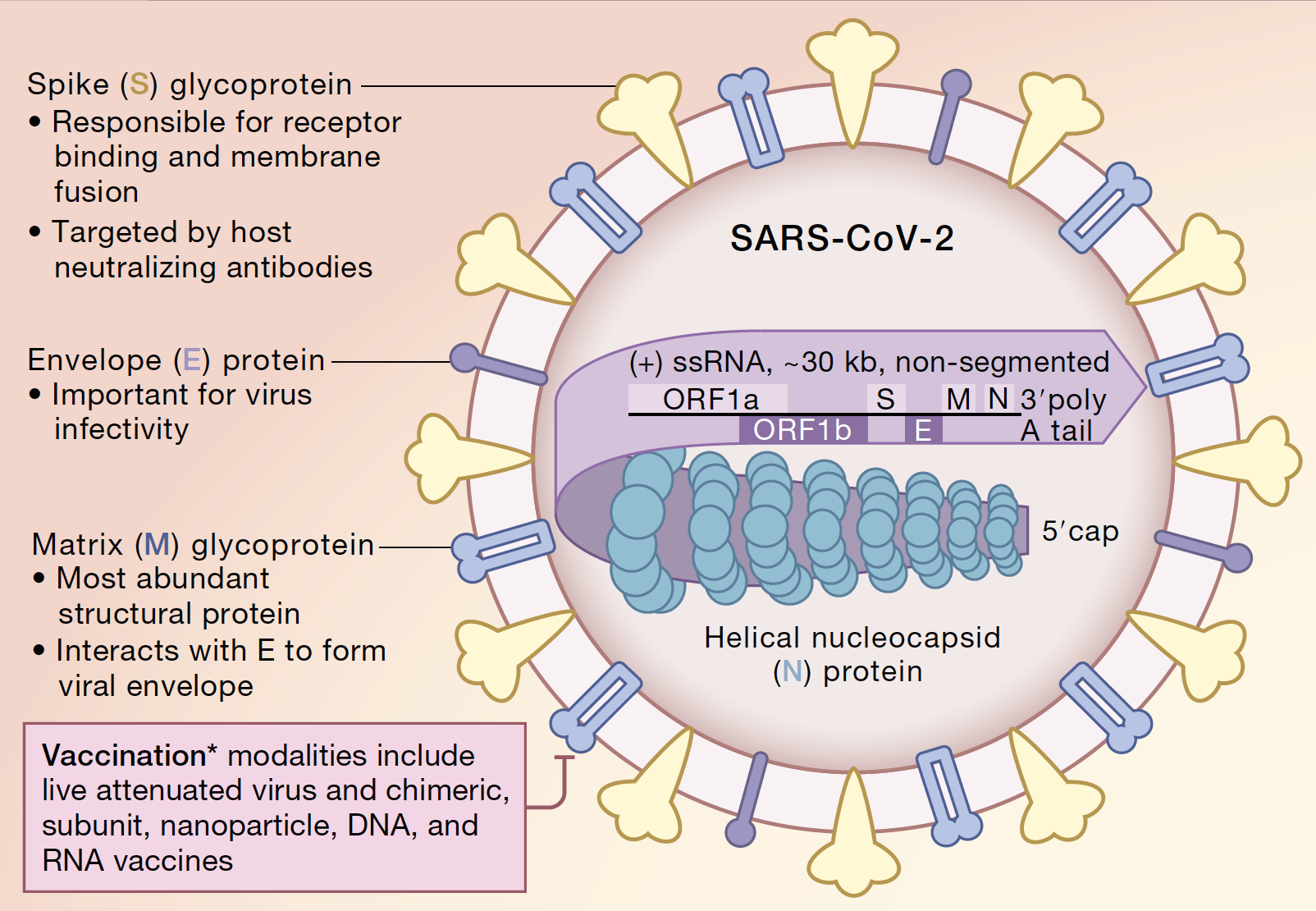
One of the most notable differences is that the genome of SARS-CoV-2 is quite large for an RNA virus (~30 kb compared to ~10 kb for most other RNA viruses). Most RNA viruses accumulate many mutations during replication due to the fact that their enzymes, which copy their RNA genome into new copies of RNA to make viral proteins and new viruses, do not have proofreading activity. SARS-CoV-2 does encode a viral protein with proofreading activity, which makes the genome of this virus slightly more stable. The genome is a plus-stranded RNA molecule that is translated by ribosomes as a single polypeptide that is eventually cleaved to multiple viral proteins by viral-encoded proteases. In addition to this method of generating viral proteins, some of the structural and accessory proteins of the virus are also translated from mRNAs generated by discontinuous transcription. The discontinuous transcription of coronavirus RNAs enables recombination between different coronavirus genomes in the same cell. It is notably unusual for a virus to use all of these mechanisms to generate proteins.
A very easy to read "Snapshot" from the journal Cell is a 2-page document with nice graphics and explanations to explain SARS-CoV-2 viral structure, life cycle, and immune response. Download it here.
Coronaviruses 101
A commendable one-hour lecture on the molecular virology of coronaviruses by Dr. Britt Glausinger from U.C. Berkeley can be found at the link below. It covers other human coronaviruses, how the viruses likely transmitted to humans from animals, genetics, replication mechanisms, and immune interactions. It is intended for a scientific audience. It explains why coronaviruses are relatively resistant to antiviral agents, like ribavirin and remdesivir, -used alone, and it describes the replication and transcription complexes that form in infected cells derived from the endoplasmic reticulum. It also discusses the delayed interferon responses to this virus that likely contribute to immunopathology-mediated disease and highlights the fact that neutralizing antibodies are often short-lived. She also highlights the need for new tractable animal models to study pathogenesis and therapeutics.
The second video is from Dr. Christiane Wobus and provides a shorter (30 minute) overview of coronavirus biology and SARS-CoV-2.
What is known about the various viral proteins?
A lucid lay summary of the functions of the various SARS-CoV-2 viral proteins is found in this New York Times article, (best viewed through Google Chrome).
Research Talks of Interest
Dr. Daniel Goldstein presents Why older people do worse from COVID-19: Lessons from Influenza in the presentation:
Research Articles of Interest
This article by Drs. Iwasaki and Yang discuss the potential dangers of vaccines that generate a suboptimal antibody response.
This article authored in part by UM's own Matt O'Meara identifies 332 high confidence SARS-CoV-2 human protein-protein interactions and identifies 66 druggable targets, including targets for 29 FDA approved drugs. https://medicine.umich.edu/dept/ebs/curated-information-covid-19
This lay article explains how genetic sequences are mapped to genetic trees and provides insight into the origins of SARS-CoV-2. It also discusses how this information disproves conspiracy theories about the virus being manufactured in a lab.
How (Not) to Do an Antibody Survey for SARS-CoV-2.
What Do Antibody Tests For SARS-CoV-2 Tell Us About Immunity?
This article authored in part by UM's own Matt O'Meara identifies 332 high confidence SARS-CoV-2 human protein-protein interactions, and identifies 66 druggable targets, including targets for 29 FDA approved drugs. Please see it here.
A new paper is Science characterized 50 COVID-19 patients to show distinct patterns of immune response. Findings showed A unique phenotype was observed in severe and critical patients, consisting of a highly impaired interferon (IFN) type I response (characterized by no IFN-β and low IFN-α production and activity), associated with a persistent blood viral load and an exacerbated inflammatory response. See the paper here.
A new study shows rhesus macaques that are reinfected with SARS-CoV-2 following recovery from a primary infection do not show detectable viral transmission or clinical manifestations of disease. These data suggest that viral infection likely can induce neutralizing immunity at least in the short term from reinfection. See the article here.
A genomewide association study involving 1980 patients with SARS-CoV-2 infection and severe disease were analyzed. In this cohort, a blood-group–specific analysis showed a higher risk in blood group A than in other blood groups (odds ratio, 1.45; 95% CI, 1.20 to 1.75; P=1.48×10−4) and a protective effect in blood group O as compared with other blood groups (odds ratio, 0.65; 95% CI, 0.53 to 0.79; P=1.06×10−5). See the whole study here.
This early study published in Cell demonstrated that adenovirus transduction of human ACE2 receptor into mice allows for mice to be infected by SARS-CoV-2 and that neutralizing antibodies could be protective
